Flow Induced Dispersion Analysis
Flow Induced Dispersion Analysis (FIDA) is a biophysical technology that measures absolute molecular size, binding interactions, and aggregation behavior directly in solution, using flow profiles and fluorescence signals.
Fida Biosystems introduces FIDA which leverages laminar flow and diffusion to monitor how a sample spreads (disperses) over time as it is passed through a capillary flow channel by a pressure gradient. From the dispersion profile, the hydrodynamic radius (Rh) of molecules is calculated with high precision. By using intrinsic fluorescence or a fluorescent tracer, FIDA can detect changes in Rh due to binding, complex formation, or aggregation. Measurements are performed in native conditions without immobilization or separation, offering a robust and quantitative readout of molecular behavior. Additionally, Fida incorporates a Lambda dynamics signal enabling small molecule protein interactions to be studied.
FIDA enables in-solution absolute sizing, binding affinity and kinetics analysis, and aggregation detection—including early oligomerization—in a single assay. It requires only minimal sample volumes and works directly in complex/unpurified matrices such as plasma, cerebrospinal fluid, or fermentation broth. The technology delivers fully quantitative, biophysics-based data with no dependency on calibration standards or assumptions. FIDA provides access to raw dispersion profiles and primary data, supporting full data transparency and facilitating integration into machine learning or AI-based analysis pipelines. Applications range from protein engineering, formulation, and quality control to biomarker studies, drug screening, and mechanistic studies of disease-relevant interactions.
FIDA provides a high-resolution, in-solution view of molecular binding and aggregation—essential for understanding dynamic interactions in biophysics, drug discovery, and translational science.
Technology
Flow Induced Dispersion Analysis (FIDA) is a proprietary solution-based, capillary technique that measures absolute hydrodynamic radius (Rh), binding interactions, and aggregation behavior, using a unique combination of flow profiles and fluorescence detection directly observing changes in diffusivity. With one assay, it derives multiple important biophysical parameters, including size, affinity, kinetics, oligomerization, molecular stability, viscosity, stickiness, and much more — without the need for immobilization or separation.
FIDA is grounded in well-established principles of physics and fluid mechanics (Taylors Dispersion and Stock-Einstein equation). A sample plug — containing either an intrinsically fluorescent analyte or a fluorescently labeled tracer — is injected into a laminar flow within a capillary. As the sample disperses, it forms a concentration-dependent profile that reflects the diffusion and flow characteristics of the molecule. This profile is directly linked to the molecule’s hydrodynamic radius. Shifts in size reflect binding events, structural changes, or aggregation. Since FIDA measures absolute size in solution, it avoids assumptions tied to molecular weight or calibration standards. It is compatible with complex matrices such as plasma, cell culture supernatants, or cerebrospinal fluid, and delivers high-resolution, quantitative data suited for both exploratory and routine workflows.
Workflow
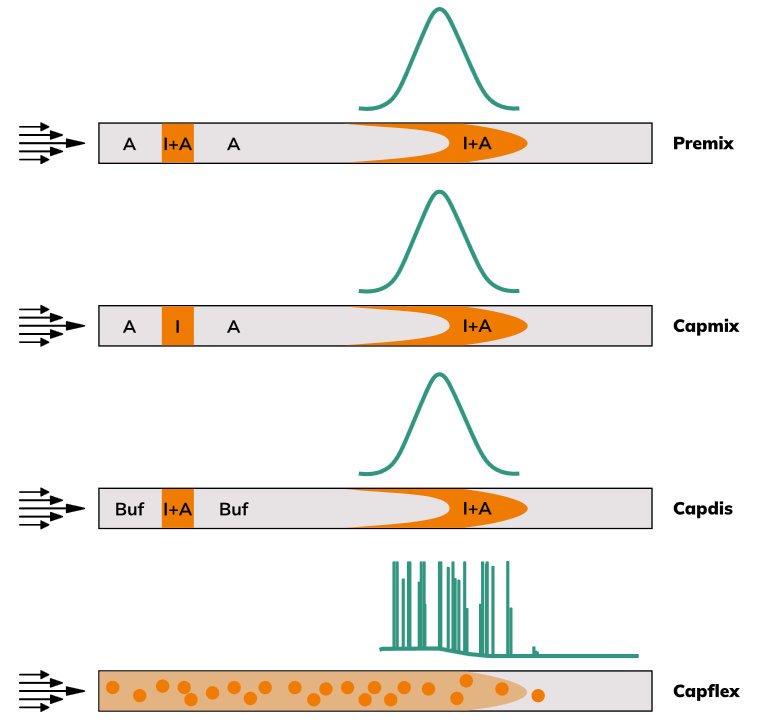
FIDA experiments are performed in solution and follow a streamlined workflow:
- An indicator—either labeled ligand or fluorescent analyte—is mixed with the sample (e.g., binding partner, complex matrix, formulation).
- The mixture is injected into a temperature-controlled capillary under laminar flow.
- The dispersion profile is recorded at the detector as the sample flows past.
- Analysis software fits the raw data to extract absolute hydrodynamic radius and Lambda Dynamics readout, which is used to generate binding curves and determine interaction constants (KD, ka and kd).
This simple assay requires only a few microliters of sample and avoids immobilization, or column separation. The measurement takes around 3 minutes per sample, and the software provides access to raw signal traces, fitted curves, and calculated parameters—ensuring full transparency and enabling integration into AI/ML pipelines or automated workflows.
Typical results
FIDA provides multiple quantitative outputs from a single experiment. The primary result is the absolute hydrodynamic radius (Rh), sensitive to changes in binding, conformational state, or aggregation. Binding affinity (KD) is determined via equilibrium shifts in Rh across concentration series. For kinetic measurements, association and dissociation rates (ka and kd) are extracted by following the dynamic change in Rh over time. In aggregation studies, Rh shifts or profile broadening reveal early oligomer formation, fibril growth, or formulation-induced aggregation. On top of that, FIDA delivers a full sample and expriment quality control overview, which includes: sample viscosity, stickiness, quantification, polydisperisty index, PDB correlator, labelling quality (free flurophore) & sample loss. All of these readouts are taken during one measurement. Results are delivered with high reproducibility and resolution, making FIDA especially useful for characterizing proteins, small molecules, or biologics under native conditions. Raw data and dispersion profiles are fully accessible, supporting advanced data analysis, publication, and regulatory documentation.
Applications
Binding affinity and kinetics in solution
FIDA allows for simultaneous determination of binding affinity (KD) and kinetic rate constants (ka and kd) in free solution, without immobilization or separation steps. This is particularly advantageous in cases where immobilization affects binding behavior or where analytes are too fragile or complex to tolerate surface interactions. The equilibrium curve is obtained from shifts in hydrodynamic radius (Rh), while kinetic parameters are derived from time-resolved measurements of complex formation. This approach has been used to study protein-protein, protein-peptide, and protein-small molecule interactions, including weak or transient binders in serum and cell lysates. For example, the measurement of antibody-antigen interactions in serum can be achieved with minimal sample and full control over stoichiometry and specificity. It is also possible to study kinetics in 100% plasma.
More on Affinity and Kinetics.
Membrane protein–ligand interaction analysis
FIDA can characterize binding to solubilized membrane proteins in nanodiscs, liposomes, or detergents—formats often incompatible with conventional surface-based biosensors. By preserving native-like conformations and working in low-concentration, detergent-rich conditions, FIDA supports high-resolution studies of GPCRs, ion channels, and other integral membrane proteins. The hydrodynamic radius readout directly reflects complex formation or conformational changes, even in partially purified samples.
More on membrane-proteins.
Aggregation and oligomerization
FIDA is sensitive to size changes associated with early-stage aggregation, including dimerization, oligomerization, and fibril elongation. Unlike techniques relying on turbidity or scattering, FIDA can resolve distinct aggregation states based on absolute size, and detect mixtures of monomer and aggregates in a single readout. The technology has been applied in the study of disease-related aggregation (e.g., α-synuclein, tau) and for monitoring protein stability under thermal or formulation stress.
More on aggregation.
Structural biology and sample screening
In protein production and structural biology workflows, FIDA provides rapid characterization of sample homogeneity, folding state, and oligomerization. This supports early selection of constructs for structural studies, where misfolded or aggregated species may compromise downstream techniques like NMR, X-ray crystallography, or cryo-EM. FIDA measurements are routinely used to screen expression conditions, purification strategies, and fusion tags, with absolute Rh serving as a proxy for conformational state and dispersion.
More on cryo-em preparation.
Characterization of lipid nanoparticles (LNPs)
FIDA is capable of detecting LNP interactions with proteins or receptors, either by monitoring size changes upon binding or by tracking aggregation. Measurements are performed in solution without dilution or labeling of the particle itself. This facilitates the study of encapsulation effects, surface adsorption, and cargo release kinetics in native-like environments. FIDA is especially suited for formulation screening or comparability assessments between LNP batches.
More on lipid nanoparticle.
Absolute size and formulation screening
By measuring hydrodynamic radius directly, FIDA can distinguish subtle changes in molecular conformation or detect formulation-induced changes in compactness or stickiness. This makes it ideal for protein formulation screening and structural studies. Unlike many methods based on molecular weight, FIDA provides size as an absolute measurement—critical for ML & AI models.
More on molecular-size.
Quantification in complex matrices
FIDA can quantify proteins, complexes, or active species in crude samples such as cell lysates, fermentation broths, or biofluids. The built-in reference standard allows for accurate concentration determination even when conventional assays fail due to matrix effects.
More on quatification.
FIDA - Instruments
Fida 1
- 3 wavelength (LED) options: 480 nm, 640 nm, 280 nm
- Size change detection of less than 1% (documented even dow to 1,3 Ångström)
- 0.5-500nm Dynamic Range
- pM-mM Affinities
Fida Neo
- All as Fida 1
- With in-solution kinetics ( both slow and fast, sec-hrs, kinetics)
- 3-fold increase in Signal-to-Noise ratio (Compared to current stateof the art detectors)
- With cSpring – a rapid, low-input method for screening protein-small molecule interactions
- Dedicated Quality Control dashboard
- See Technical specs on this product
FIDA - Suppliers
Fida Biosystems ApS
Fida Biosystems ApS
Generatorvej 6A
2860 Søborg
Denmark
Tel: +45 28 19 25 80
website:
www.fidabio.com
e-mail :